1. I. Introduction
he current theory on Attention Deficit Hyperactivity Disorder (ADHD) emphasizes on delayed maturation of brain regions involved in controlling executive function (EF), 1,2 thus leading to ageinappropriate impulsivity, hyperactivity and inattention. 3 Though deficit in inhibitory control mechanisms was earlier hypothesized as the major cause for improper EF, 4 recent studies revealed that this is primarily moderated by deficits in basic information processing. 5 Apart from the core symptoms, individuals with ADHD frequently suffer from co-morbid learning difficulty (LD), oppositional defiant disorder, and conduct disorder, 3 which also could be due to improper information management.
Image analysis revealed significant reduction in the prefrontal cortex (PFC) volume of ADHD probands. 6 As PFC is interconnected with other brain regions like the neocortical regions, amygdala, limbic circuit and cerebellum, it was proposed to have vital role in memory encoding and retrieval as well as decision making, 7 emotion related arousal, 8 and motor movements. 9 PFC microcircuits are supposed to play key roles in perception of action cycle while dealing with different types of environmental and social stimuli thereby executing a particular behavioral response. 10 Thus sustained attention and information processing, mediators of executive processes, may also be regulated by PFC.
As proposed by Dr. Barkley, EF involves six sets of self regulatory activities, such as self-inhibition, selfdirected sensory-motor action, self-directed private speech, self-directed emotion/motivation, self-directed play, and self-monitoring which eventually affect future consequences. 11 He also concluded that these six functions form the Instrumental-Self-directed level of EF that is most proximal to PFC development and functioning. Self inhibition, spatial management and sustenance of self-motivation form part of these selfregulatory behaviors and injuries / or developmental anomalies of the PFC were found to disturb these functions. Dopamine (DA) is one of the major neurotransmitter involved in movement, motivation and other executive processes 12 and the PFC is enriched with DA receptors, both type I and II. While bioavailability of DA in the PFC and striatum is regulated by DA receptor 2 (DRD2), receptor 4 (DRD4) and DA transporter, 13 PFC is preferentially enriched with DRD4. 14 A dual role of DRD4 on ?-amino-3-hydroxy-5-methyl-4isoxazolepropionic acid (AMPA) receptor, hypothesized to underlie the mechanisms of evoke related response, inhibitory control and other cognitive processes, has also been documented; during the hyper-activated state of the PFC, DRD4 was found to reduce glutamatergic transmission while at the hypoactive state PFC was reported to trigger AMPA response via the same pathway. 15 Genetic polymorphisms in the DRD4 have been explored widely. The most frequently investigated site is a variable number of tandem repeat in the exon3 and meta-analysis revealed association of higher repeats (>6R) with ADHD in the Caucasoid 16 as well as Indo-Caucasoid probands. 17 Individuals homozygous for the common 4R variant showed reduction in the PFC gray matter volume 18 while being less efficient in a measure of executive attention. 19 Based on these findings, we speculated that DRD4 may have a contributory role in the EF of ADHD probands and for the first time investigated association of functional DRD4 promoter variants with Erratic Organizational Capability (EOC) and Short Attention sustainability (SAS), as part of selfregulatory trajectories under EF, in eastern Indian probands with or without co-morbid LD.
2. II. Methods
3. a) Participants and study design
Nuclear families with ADHD probands (N=200; mean age 7.7 yrs; sex ratio M:F 9.5:1) were enrolled based on the Diagnostic and Statistical Manual for Mental Disorders-IV-text revised (DSM-IV-TR) criteria. 3 ADHD index, hyperactivity level and cognitive attributes/ inattentiveness of probands were measured by the Conners' Parent Rating Scale-Revised (CPRS). 20 Intelligence/developmental quotient were assessed by the Wechsler's Intelligence Scale for children 21 for proband above five years and Developmental Screening Test for children below 5 years. 22 Out of 200 probands, 160 were complete parent-proband trios, 22 had only one parent while 18 were affected probands only. Majority of the probands belonged to the combined subtype (72.5%) while hyperactive/impulsive (12.5%) and inattentive (15%) subtypes were only few. 60% probands showed cognitive deficit while 63% exhibited hyperactivity. Co-morbid conditions assessed using the DSM-IV-TR criteria 3 showed LD in 29% of probands. Subjects with only psychiatric problems including pervasive developmental disorders, any form of mental retardation (IQ ? 70) and fragile-X syndrome, were excluded.
Ethnically matched control subjects were evaluated for the DSM-IV-TR criteria for ADHD 3 , hypothyroidism, intelligence/developmental quotient (>80) as well as for any psychiatric disorder running in the family and those without any abnormality (N=200; Mean age 13.45 yrs; sex ratio 1.4:1) were recruited. Informed written consent was obtained for participation in the study and the protocol was approved by the Institutional Human Ethics Committee.
4. b) Assessment of traits
SAS and EOC were measured through questions selected from the DSM-IV-TR and CPRS scale (Table 1), scores (0-3) were given based on the responses received, and the total score was converted to percentage. CPRS score percentage for each trait was cross validated with the DSM-IV-TR score and an individual exhibiting more than 5% deviation was excluded. Based on the CPRS score percentage, individuals having less than 30% were considered to have low deficit (1), whereas those with 30-60% were identified as having medium deficit (2) and more than 60% were coined as having maximum deficit (3).
Computerized games were used to measure the cognitive function of ADHD probands (N=25, 6-12 yrs, 23 male / 2 female) and controls (N=10, 7-12 yrs, 6 male / 4 female). Participants were tested for working memory (Game 1), speed (Game 2) and spatial (Game 3) information processing, , and dual N back test (Game 4) for 5 minutes. For each game, there were 3 levels with increasing complexity followed by automatic recording of score. Wrong entry in any round iterated the same level and thus score increased with delay/error in response.
Probands (N=80; mean age 12.67 ±3.95 years) were reassessed after 3 years using the same questionnaire (Table 1) to follow their performance.
5. c) Genetic analysis
Online programs F-SNP (compbio.cs.queensu. ca/F-SNP/), Brain-array (http://brainarray.mbni.med. umich.edu/brainarray/database/searchsnp/snpfunc.asp x), and SNPinfo (http://snpinfo.niehs.nih.gov/cgi-bin/ snpinfo/snpfunc.cgi) were used to analyze functional roles of seven upstream variants. Peripheral blood leukocytes were processed for extraction of genomic DNA. 23 Oligonucleotides designed using the Primer3 (www.bioinformatics.nl/primer3plus/) program were used for PCR amplification in ABI Gene Amplifier #9700 PCR system. rs 916455 was genotyped by restriction fragment length polymorphism analysis of PCR amplicon using RsaI restriction enzyme (New England Biolab); in presence of the "T" allele, two fragments of 58 and 163 bp were generated. The other SNPs were analyzed by sequencing of the PCR amplicon in Applied Biosystems 3130 Genetic analyzer using Big Dye v 3.1 chemistry and Sequencing Analysis Software, v 5.2.
6. d) Data analysis i. Association analysis
Unphased verion 3.1.7 24 was used for populationand family-based analysis. Hardy-Weinberg equilibrium (HWE) was analyzed using the online software (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl-hwe) and Piface version 1.72 25 was used to quantify the strength of statistically significant results (P<=0.05). The Odd's ratio (OR) was calculated by online program (http://www.hutchon.net/ConfidOR.htm).
7. ii. Analysis of interaction between the sites
Interaction between haplotypes was analyzed by the Cocaphase program. Linkage Disequilibrium was calculated using the Haploview program. 26 SNP-SNP interaction was analyzed by the Multifactor dimensionality reduction (MDR) program. 27 Volume XVI Issue II Version I
Year 2016 ( D D D D ) A iii.8. Genotype-phenotype correlation analysis
Association between each phenotypic trait and the gene variants were analyzed by Mann-Whitney test (http://elegans.som.vcu.edu/~leon/stats/utest.html).
Association between genotypes and co-morbid LD was analyzed using the Cocaphase program.
ADHD probands were grouped into three categories, all cases, ADHD with co-morbid LD (ADHD+LD) and without LD (ADHD-LD) for analyzing the level of SAS and EOC. Frequency of probands having various levels of SAS and EOC, calculated through CPRS, were analyzed using the excel work book. Correlation between pair of traits was obtained through online Pearson's calculator (http://www. socscistatistics.com/tests/pearson/) and regression analysis software (http://www.alcula.com/calculators/ statistics/linear-regression/) was used for calculating the interdependence of these traits.
9. iv. Measurement of cognitive function
Mean scores obtained for ADHD probands and controls through computerized assessment were analyzed by the 1 tailed unpaired T test using online software (http://studentsttest.com/).
10. III. Results
11. a) Analysis of variants
Sequence analysis showed presence of a novel G>T substitution (Table 2, NSNP) 45 bases before rs747302. All the seven SNPs are binding sites for transcription factors and four revealed moderate regulatory potential (Suppl Table S1). rs916455 is located in the CpG island (ratio=0.99).
Genotypes of rs747303 deviated from the HWE in the probands (P=0.0009). rs10902180 genotypes deviated for the proband (P=0.0001) as well their parents. Genotypes of all other variants followed the HWE. Population based analysis showed significant bias for rs10902180 "C" allele (Suppl Table S1; P=0.01, Power=71, OR=1.57) with a trend of association (P= 0.08) for the "CC" genotype (Table 2). rs916455 "CC" and rs936462 "AA" genotypes showed significantly higher frequencies in the ADHD+LD probands as compared to control as well as ADHD-LD (Table 2, P<0.04). rs10902180 showed higher frequency of the "GG" genotype in the ADHD+LD compared to ADHD-LD individuals (P=0.001).
Family-based analysis revealed biased transmission of rs936462 "A", rs747303 "T", rs1800955 "T" and NSNP "G" alleles (Table 3). For rs1800955 "T", a paternal bias was noticed (? 2 = 6.32, P=0.01). Analysis of haplotypes failed to show any significant difference.
Linkage Disequilibrium pattern was different in the control individuals, probands and their parents, but coefficient of correlation was insignificant (Suppl Fig. 1).
12. b) Analysis of phenotypic traits
EOC and SAS showed linear correlation in both ADHD-LD (R=0.73) and ADHD+LD (R=0.88). Regression analysis validated EOC score as a function of SAS for these subgroups (Y= 2.31+0.86X & y= 1.08X-10.30 respectively). Analysis between different subgroups exhibited higher number of ADHD+LD probands with high SAS score (? 2 =21; p>0.0001) as compared to ADHD-LD group (Suppl Fig. 2). No significant difference was noticed for the EOC score (Suppl Fig. 2). rs747303 "TT" showed association with higher EOC score (Suppl Table S2, P=0.05), while the NSNP "GG" showed association with both high EOC and SAS scores (P=0.02 & 0.04 respectively).
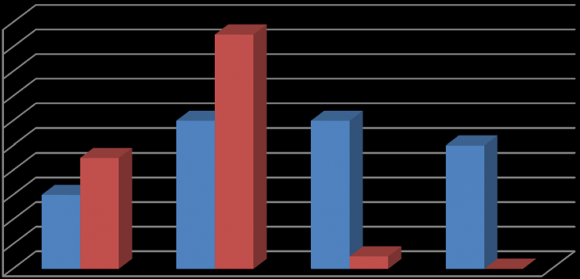
Performance of ADHD probands was poor, more strikingly for Game 1 and 2, as compared to agematched control children (Fig. 1).
Association of higher scores for Game 1 and 2 with rs916455 "CC" was observed. rs936462 "AA" and rs747303 "TT" revealed nominal differences, while rs10902180 "GC" showed distinct difference in Game 2 score with a mild difference for Game 1 (Suppl Fig. 3). Higher mean score was also noticed for rs1800955 "CC" in case of Game4. No difference could be observed for rs747302 and data for NSNP could not be shown due to the presence of only one heterozygote (Suppl Fig. 3).
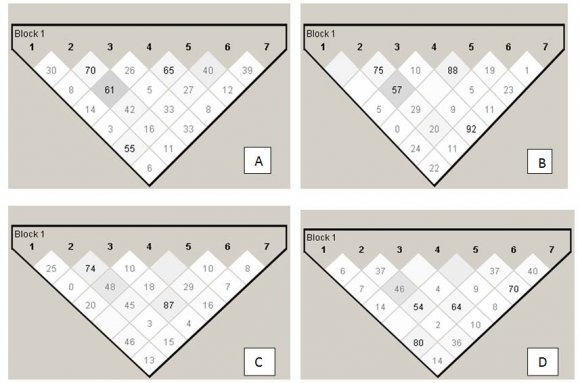
Interaction analysis revealed major independent effects of both SAS and EOC (Fig. 2 A & B respectively) in ADHD individuals exhibiting higher scores (score>1) against those having low score (score=1). With EOC as a phenotypic co-variate, interaction between rs916455-rs747302 and rs1800955-NSNP was also noticed (Fig. 2B). Stratification based on the presence of co-morbid LD revealed major independent effects of phenotypic traits and gene variants in ADHD-LD probands as compared to the control individuals (Fig. 2C), while in ADHD+LD individuals, strong interactive effect was observed between SAS-rs1800955 and EOC-rs747303 in absence of any major independent effect (Fig. 2 D), as compared to ADHD-LD individuals. Mild positive interaction was also noticed between SAS-rs747302, SAS-rs747303, and EOC-rs1800955 (Fig. 2D).
Follow up after three years showed that while the number of probands with high EOC gradually reduced with time (Suppl Fig. 4, Low T0/T3=2/24, High T0/T3= 44/15, ?2=32.5, P=0.0001), SAS score improved in a number of probands (Suppl Fig. 4, Low T0/T3= 0/11, High T0/T3= 56/48, ?2=11.5, P=0.003). ADHD subjects harboring rs916455CC, rs747303TT and NSNPGG genotypes had higher EOC scores after three years, while NSNP also showed association with high EOC score (Suppl Table S3). Follow up study also revealed strikingly low scholastic improvement in ADHD+LD (58%) probands as compared to ADHD-LD (79%).
13. IV. Discussion
Earlier investigators reported delayed maturation of brain regions controlling EF, affecting self regulation, attention and working memory. 2 Since these regions are enriched with DRD4 receptor, we investigated association between DRD4 promoter variants and EF of ADHD probands. LD is a major comorbid condition and may result from low attention sustainability, memory retrieval, working memory, and poor comprehension. We compared the genotypic pattern of ADHD+LD individuals with that of ADHD-LD individuals as well as controls to find out if any particular genotype is affecting the trait.
Based on the data obtained, we for the first time report significant association of DRD4 promoter variants with EF deficit of Indo-Caucasoid ADHD probands. F-SNP analysis revealed that rs9164555, an upstream variant, may regulate binding of transcription factor, though the mechanism is yet to be understood. The rs916455 "C" allele showed association with persistence of symptoms in Chinese ADHD subjects. 28 Follow up of ADHD probands during the present study also revealed association of "CC" with high EOC score. Higher occurrence of the "CC" genotype was earlier reported in ADHD+LD probands 29 and further analysis in extended samples also revealed association of the "CC" genotype with ADHD+LD as compared to controls (P=0.05) as well as ADHD-LD (P=0.04). MDR analysis exhibited additive effect of rs916455 and rs747302 on EOC. In ADHD+LD individuals, this site showed strong independent effect. "CC" was also associated with Game 1 and 2, depicting its role in working memory impairment as well as poor cognitive flexibility while follow up revealed link between the "CC" genotype and poor attention.
rs747302, presented as a trimorphic variant (C/A/G) in the dbSNP database (build 86/142), showed only two alleles (C/G) in the present study as well as previous investigations. 30, 31 F-SNP analysis suggested that the C allele affects binding of transcription factor E2F. Comparative analysis failed to show any significant association of rs747302 with ADHD in the Indo-Caucasoid population and further investigation in other ethnic population is warranted to understand the actual role.
Frequency of rs936462 "A" allele was 50% less in the studied Indo-Caucasoid population as compared to the Hungarian population. 31 We have noticed preferential transmission of the "A" allele by familybased analysis (Odds ratio 4.73). Individuals harboring "AA" showed higher score for Game 1, 3 and 4. While Game 1 is a test for working memory, Game 3 and 4 requires sustained attention and organizational efficiency. Therefore, the "A" allele may be considered as a risk allele in this population. A previous report showed that absence of the "G" allele caused a significant difference in the genotype of -521 C/T, i.e. rs1800955, 31 though in the studied Indian population no such difference was noticed. On the basis of the present study, rs936462 merits further analysis to understand the role of the site in the disease etiology.
rs747303 was rarely investigated in ADHD patients and the present study revealed biased transmission of the "T" allele (OR 2.81). F-SNP analyses suggested regulation of transcriptional activity; the GC box disappears in presence of the T allele thus affecting transcription initiation. ADHD probands with the "TT" genotype had poorer information processing capability as compared to probands harboring the "GG" genotype. This is also indicated by scores for Game 3 & 4. MDR analysis showed positive effect of this site on poor attention span in ADHD+LD subjects. On the basis of these findings, rs747303 "T" allele could be considered as a risk variant for ADHD which merits further in depth analysis.
This first association analysis on rs10902180 identified the site as a transcriptional regulator. Marginally higher frequency of the "C" allele and "CC" genotype was noticed in the ADHD probands as compared to control. Individuals with the "CC" genotype obtained higher scores for Game 1 and 2. On the other hand, analysis among the subgroups showed significantly higher frequency of the "GG" genotype in ADHD+LD. This contradictory finding may suggest a different mechanism of DRD4 expression in the ADHD+LD subgroup since the gene not only interferes with NMDA receptor or other D2 type receptors, but also interacts with D1 type D5 receptors which work by upstream regulation of gene (analyzed by KEGG pathway). MDR analysis showed strong independent effect of this site on the phenotypic traits. Since our study involves only limited number of ADHD+LD probands, we conclude that this site may have a role in the learning problem of ADHD probands which merits further analysis in higher number of subjects.
rs1800955 is a transcriptional regulator widely investigated in ADHD as well as other psychiatric disorders. 32 Transcription factor CAP is functional in presence of the "T" allele; transcriptional activity was reduced by 40% in presence of the "T" allele 33 though the finding could not be reproduced. 34 Earlier studies on the Indo-Caucasoid population revealed biased parental transmission of haplotype 7R-T of DRD4 Exon3 VNTR and rs1800955. 35 The present study also revealed parental over transmission of "T" to the probands, which is basically paternal in nature. "CC" was associated with higher scores for SAS and EOC measured through CRS as well as Game 2 and 4 suggesting its role in cognitive impairment as a whole. MDR showed strong independent effect of rs1800955 in ADHD. In ADHD+LD both attention sustainability and information processing was found to be affected in presence of this variant The novel substitution NSNP detected in the 5' upstream region showed a parental bias in transmission of the wild type allele and interaction with rs1800955. The heterozygous form showed association with both SAS and EOC. However the site failed to show any significant functional contribution thus making it difficult to interpret its role.
Linkage Disequilibrium between rs747302-rs1800955 and rs916455-rs1800955 in the Indo-Caucasoid control population was similar to that observed in the Japanese population. 36 However, in the Hungarian population, a strong bond was noticed between rs936462-rs1800955 30 which was absent in the Indian population. Further, in families with ADHD probands, the pattern was totally different as compared to the ethnically matched control population. From the observed pattern, we may interpret that the DRD4 promoter region harbors recombination hotspots which culminates in a break in the Indo-Caucasoid population.
ADHD associated EF deficit was hypothesized to occur from poor flexibility, self motivation and working memory, ultimately giving rise to altered behavioral response. 11,37 Uncontrolled inhibition with triggered impulsivity and error prone behavior was also noticed. 37 Further investigation showed improper information processing as the major reason for ADHD associated symptoms. 5 These domains are supposed to be affected in children with LD too. As In the present study, we have noticed aberrant information processing along with short attention sustainability. Higher scores for Game 1 and 2 in ADHD probands indicate poor working memory and cognitive flexibility as a result of improper information processing. Scores for Game 3 and 4 were moderately high in the ADHD probands as well as healthy individuals which may indicate that these traits involve a more complicated network of information processing which develops during adolescence. MDR analysis also revealed strong major effects of these two phenotypic traits in addition to independent effect of the studied sites and an additive effect of rs916455-rs747302 on EOC. Comparative analysis between subgroups showed that phenotypic traits of ADHD+LD subjects are affected more severely by interactive effect of the markers; while in ADHD-LD both SAS and EOC showed strong independent effects, interactive affects were pronounced in ADHD+LD. Follow up revealed a constant deficit in attention sustainability with a gradual improvement in EOC and academic achievement was worse for ADHD+LD patients. rs916455 "CC", rs747302CC, rs936462 "AA", rs747303 "TT", rs1800955 "CT/TT" and NSNP "GG" were found to be more frequent in subjects with high and medium score for SAS and EOC indicating significant impact of these genotypes in the cognitive function. Follow up study also confirmed role of rs747303 "TT", rs1800955 "TT" and NSNP "GG" in ADHD.
Since ADHD probands are believed to have an altered function of the frontal lobe 38 and DRD4 density is high in this region, we speculated that the promoter variants may alter transcriptional activity leading to a reduction in DRD4 receptor density, thereby causing altered behavioral and cognitive outcome. The data obtained indicate that failure in information processing, leading to reduction in attention span, may lead to the symptoms of ADHD which is more evident in subjects with co-morbid LD. Further analysis involving additional functional variants is warranted in large cohort of subjects to validate our observation. Supplementary Figure 1 : LD analysis for all ADHD probands (A), Father of the probands (B), Mother of the probands (C), ethnically matched healthy individuals (D).
14. Volume XVI Issue II Version I
15. Legend to Figures
Volume
| ID | Genotype | Control (N=200) | All (N=200) | ? 2 (P) | Probands (N=146) (N=54) ADHD-LD ADHD+LD | ? 2 (P)* | ? 2 (P)! | |
| rs916455 | CC | 0.88 | 0.89 | 1.53 | 0.85 | 0.94 | 6.34 | 6.34 |
| CT | 0.12 | 0.10 | (0.47) | 0.14 | 0.04 | (0.05) | (0.04) | |
| TT | 0.0 | 0.01 | 0.01 | 0.02 | ||||
| rs747302 | CC | 0.34 | 0.39 | 0.95 | 0.37 | 0.41 | 1.22 | 1.82 |
| GC | 0.46 | 0.43 | (0.62) | 0.46 | 0.37 | (0.54) | (0.40) | |
| GG | 0.20 | 0.18 | 0.17 | 0.22 | ||||
| AA | 0.90 | 0.93 | 3.22 | 0.91 | 1.0 | 7.43 | 9.42 | |
| rs936462 | GA | 0.09 | 0.07 | (0.20) | 0.09 | 0.0 | (0.02) | (0.002) |
| GG | 0.01 | 0.0 | 0.0 | 0.0 | ||||
| rs747303 | GG | 0.09 | 0.10 | 3.97 | 0.10 | 0.12 | 4.44 | 1.79 |
| GT | 0.36 | 0.26 | (0.14) | 0.28 | 0.20 | (0.10) | (0.41) | |
| TT | 0.55 | 0.64 | 0.62 | 0.68 | ||||
| GG | 0.77 | 0.66 | 5.02 | 0.61 | 0.83 | 1.06 | 13.2 | |
| rs10902180 | GC | 0.17 | 0.24 | (0.08) | 0.26 | 0.14 | (0.59) | (0.001) |
| CC | 0.06 | 0.10 | 0.13 | 0.03 | ||||
| rs1800955 | CC | 0.19 | 0.16 | 0.93 | 0.16 | 0.15 | 2.08 | 0.78 |
| CT | 0.51 | 0.50 | (0.62) | 0.49 | 0.44 | (0.35) | (0.68) | |
| TT | 0.30 | 0.34 | 0.35 | 0.41 | ||||
| NSNP | GG | 0.82 | 0.80 | 0.10 | 0.81 | 0.77 | 0.47 | 0.48 |
| GT | 0.18 | 0.20 | (0.74) | 0.19 | 0.23 | (0.49) | (0.49) | |
| TT | 0.0 | 0.0 | 0.00 | 0.00 | ||||
| SNP | Allele | Transmitted | Not transmitted | ? 2 (P) | Power (%) Odds Ratio | |
| rs916455 | C | 0.96 | 0.94 | 1.47 | --- | --- |
| T | 0.04 | 0.06 | (0.23) | |||
| rs747302 | C | 0.63 | 0.59 | 0.64 | --- | --- |
| G | 0.37 | 0.41 | (0.42) | |||
| rs936462 | A | 0.99 | 0.93 | 9.01 | 85 | 4.73 |
| G | 0.01 | 0.07 | (0.003) | (1.15-19.41) | ||
| rs747303 | G | 0.10 | 0.25 | 17.7 | 99 | 2.81 |
| T | 0.90 | 0.75 | (2.59e-005) | (1.36-5.82) | ||
| rs10902180 | G | 0.83 | 0.82 | 0.07 | --- | --- |
| C | 0.17 | 0.18 | (0.79) | |||
| rs1800955 | C | 0.37 | 0.48 | 5.54 | 65 | 1.52 |
| T | 0.63 | 0.52 | (0.02) | (0.89-2.74) | ||
| NSNP | G | 0.97 | 0.82 | 27.15 | 99 | 4.89 |
| T | 0.03 | 0.18 | (1.89e-007) | ( 1.94 -12.06) | ||
| Supplementary | ||||||
| Supplementary |
| Score for phenotypic traits | ||||||||||
| ID | Genotypes | SAS | EOC | |||||||
| Low | Medium | High Low | Medium | High | ||||||
| rs916455 | CC CT | 100 0 | 100 0 | 92 8 | 90 10 | 93 7 | 100 0 | |||
| CC | 13 | 64 | 46 | 32 | 52 | 64 | ||||
| rs747302 | GC | 62 | 29 | 36 | 50 | 28 | 36 | |||
| GG | 25 | 7 | 18 | 18 | 20 | 0 | ||||
| rs936462 | AA GA | 89 11 | 100 0 | 93 7 | 91 9 | 96 4 | 93 7 | |||
| GG | 22 | 0 | 7 | 14 | 4 | 7 | ||||
| Year 2016 | rs747303 rs10902180 | GT TT GG GC | 22 56 78 22 | 8 92 50 29 | 28 65 77 14 | 36 50 73 23 | 17 79 74 10 | 14 79 79 7 | ||
| CC | 0 | 21 | 9 | 4 | 16 | 14 | ||||
| CC | 12 | 7 | 12 | 10 | 16 | 7 | ||||
| Volume XVI Issue II Version I | ID rs916455 rs747302 rs936462 rs747303 rs1800955 NSNP | Predicted functional score Allele F-SNP SNPinfo 0.109 0.148029 C T 0.208 0.086621 C G 0 0.086409 A G 0.208 0.172765 G CT 44 57 TT 44 36 GG 63 83 GT 37 17 | 49 39 74 26 | Frequency Control Probands 0.942 0.941 0.057 0.058 0.58 0.61 0.42 0.39 0.95 0.97 0.05 0.03 0.27 0.23 45 26 45 26 63 82 37 18 | ? 2 (P) 0.0002 (0.96) 0.78 (0.37) 1.62 (0.20) 1.67 57 57 77 23 | |||||
| T | 0.73 | 0.77 | (0.20) | |||||||
| D D D D ) | rs10902180 | 0.05 | 0.163562 | G | 0.86 | 0.78 | 6.39 | |||
| ( A | C | 0.14 | 0.20 | (0.01) | ||||||
| rs1800955 | 0.176 | 0.181188 | C | 0.45 | 0.41 | 0.91 | ||||
| T | 0.55 | 0.59 | (0.34) | |||||||
| NSNP | None detected | -- | G | 0.91 | 0.90 | 1.00 | ||||
| T | 0.09 | 0.10 | (0.75) | |||||||
| ID | Genotype | EOC | SAS | |||||||
| Mean± SE | P value Mean± SE | P value | ||||||||
| rs916455 | CC | 65.38 2.06 | -- | 70.51,1.66 | -- | |||||
| CT | 65.19 5.34 | 71.82, 3.96 | ||||||||
| rs747302 | CC | 66.72 3.13 | 74.37,2.24 | -- | ||||||
| GC | 66.41 2.77 | -- | 68.04, 2.61 | |||||||
| GG | 60.99 5.08 | 71.05, 3.79 | ||||||||
| rs936462 | AA | 65.17 2.02 | -- | 71.17,1.66 | -- | |||||
| GA | 69.67 6.96 | 67.60, 6.31 | ||||||||
| rs747303 | GG | 58.78±4.43 | -- | 71.71±4.44 | --- | |||||
| GT | 64.90±3.90 | 70.06,3.35 | ||||||||
| TT | 67.10±2.53 | 0.05 | 71.59±1.99 | |||||||
| rs10902180 | GG | 67.21±2.20 | -- | 70.21,1.89 | -- | |||||
| GC | 61.91± 4.83 | 73.68,3.91 | ||||||||
| CC | 61.46±6.51 | 72.67,4.39 | ||||||||
| rs1800955 | TT | 68.40±3.11 | 0.09 | 72.7±2.75 | -- | |||||
| CT | 62.51±2.73 | 69.31±2.19 | ||||||||
| CC | 69.30±5.21 | -- | 74.68,4.13 | |||||||
| NSNP | GG | 67.48±2.15 | 0.02 | 72.37±1.81 | 0.04 | |||||
| GT | 56.92±4.78 | 64.67±4.01 | ||||||||
| Year 2016 |
| D D D D ) |
| ( A |