1.
Opinion opological DNA assemblies governed biological processes, physical manipulations, compartmentalization, transfer genetic information by sequence, participates in molecular mechanisms of formation and deformation of biological processes, and even chromosome territory formation, including replication, transcription, and gene regulation via dynamic assets and variations. [1] The sequencing processes deal with DNA mechanical code and leave impacts on gene regulation via nucleosome positioning. Therefore, a proper analysis could predict the mechanical route of DNA sequencing and how it does influence loop creation. The earlier mentioned predictions testified via in-vivo transcription and in-vitro single-molecule assays. These styles of elucidation based on theoretical investigations of various cellular routes of biological processes such as sequencedependent features of DNA, a specific sequence creation in the chromatin structure, and interfaces of protein and DNA molecules. As referred before, the nonspecific protein-DNA interactions are a significant feature of dynamics paths involved in DNA loop formations. [2] Alternatively, to have a proper understanding of interactions and conformational dynamics, a more scientific analysis of molecular simulations will defiantly offer experimental and theoretical insight. The phenomenon of DNA looping participates in biological processes, including transcription, recombination, and replication, as well as gene regulation, recombination, and chromosomal activities. The structural features and physical interactions of proteins are the strategic factors associated with DNA looping. [3] These biophysical individualities can change the length scale transforms during looping. Changes in the conformation and mechanical deformation of the DNA initiate thermal fluctuations and govern the thermodynamics by generating entropy to influence looping and unlooping processes.
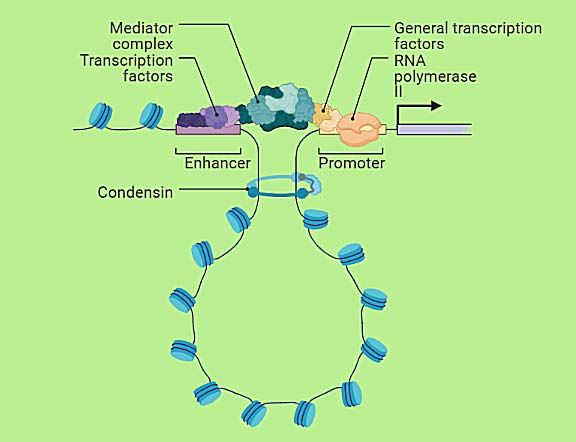
A theoretical model was prescribed, which explained the protein interactions, DNA mechanics, and conformational entropy-defined DNA looping and unlooping, and reply unanswered queries such a show this phenomenon does affect it. These insights proved that DNA deformation and entropy affect the kinetics of the looping and unlooping process. [4] Deformability, bendability, and variability of the DNA chain tempers the kinetics and disturb the interaction. The biophysical and thermodynamics change aloofness in earlier scale predictions. These perturbations and conformational changes manipulate genetic information. An experimental and theoretical insight dreadfully appropriate for the systematic quantitative divisions, and further, it answers back the queries, such as how DNA sequence does disturb looping at such a scale. Few transcription factors such as concentration, length, and sequence influence the phenomenon of DNA looping. These changes can be calculated experimentally and theoretically for better insight into the distinctions between mechanics of nucleosome formation and looping in the short length scale (figure 1). [5] These derived mechanisms of DNA-looping are too important for different types of machinery of networks of DNA metabolism, including transcription, recombination, and replication. The routes of replication and transcription types of machinery are too complicated, and therefore a better elucidation of these mechanics can expose genomic instability by providing a better clarification of transcription-replication collision mechanisms. The experimental and theoretical analyses of these biological and biophysical processes help specify the encounters that set off via transcription-replication and support co-orientation of replication and transcription. [6] These discoveries are pathfinder and a source of scientific events that show directions on how to avoid or resolve transcription-replication collisions, for example, alterations in DNA supercoiling hindering replication or chromatin-remodeling complexes.
Transcriptionreplication and DNA damage intertwined at the collisions. At once, some types of machinery of transcription-DNA replication encounter each other at regular intervals and originate genome instability to promote diseases. [7] Several factors and mechanisms exist in cellular types of machinery for inhibiting, blocking, or resolving these unusual events of cell physiology.
Further, the transcription backings mitotic recombination that will have replicate fork progression, provoking its evading and breaking. [8] In simple words, this phenomenon can address as cross-talk between transcription and recombination, in which originated conflict initiates recombinogenic DNA breaking and cotranscriptional R-loops formed. [6]One of the authors, (MB) identified the aforesaid occurrence as one of the major causes of DNA genetic reshuffling. Further, he stressed that these newly originated interfering events occurred between transcription and replication. A few queries emerged from it, such as "does it have similarities with the route of genome dynamics influenced by RNA." In this opinion, some other similar emerging questions and outlooks are discussed based on the interference between transcription and replication, as well as the way RNA influences genome dynamics. Another author (RK) pointed out Gquadruplexes that governed transcription, translation, and immunoglobulin gene reshuffling. Both agreed and marked that one cellular event as earlier chatted is hindering DNA replication machinery by these guaninerich assemblies as a piece of evidence. Recently published research articles covered the role of natural strategies such as homologous recombination and exclusion of edifice by helicases, which can pass Gquadruplex-mediated replication obstacle. [9] Such experimental and theoretical insight further provide fundamental intuitions on the routes monitoring practice when DNA looping is initiating pieces of machinery of transcription, recombination, and replication.
The mechanism of DNA loop formation is crucial and plays its role in many cellular mechanisms in different and adverse conditions. The above-stated routes are participating in governing cellular processes properly and can influence these routes of protein synthesis according to the needs. [10] The distance between the binding sites is aparameter and can affect the ability of DNA to form loops. The conformation of a particular sequence and other concerning features affect the deformability and bendability. [11] The cellular metabolic or environmental circumstances exaggerated by the extra-or intracellular signals and directly influence DNA loops. The site-specific protein-DNA binding is another phenomenon that deals with the protein-protein and protein-ligand interactions. [12] These biophysical interactions originate from different physiological states and influence many cellular processes, including B transcription, recombination, and replication. [13] Various biological molecules were applied during distinct binding topologies and in hyper-stable or hypostable loops for altering conformations. It is a wellknown fact that the phenomenon of DNA looping alters looping behaviors, cell-to-cell variability, topologies, and earlier described interactions. [14] Here, author assumed that the mechanism of loop switching can useful in controlling gene expression experimentally.